
The CIR team presented a wealth of new results at the MICCAI'25 and its connected workshops in the area of machine learning and medical image analysis ranging from brain development, cancer imaging, and the detection of novel diseases.
Predicting treatment dynamics in breast cancer
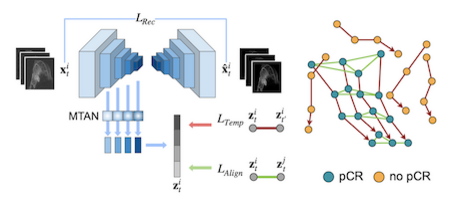
This work by Ivana Janisckova and colleagues proposes a model that learns treatment response patterns from longitudinal breast MRI scans to predict pathological complete response (pCR) in patients undergoing neoadjuvant chemotherapy (NACT). By constructing trajectories in a representation space, the method captures patient-specific temporal dynamics, aligns trajectories of complete responders to extract population-level response patterns, and takes into account the heterogeneity of response types within the non-responder group. Experiments demonstrate that this approach surpasses state-of-the-art performance in predicting NACT response. It is part of the PREDICTOME project.
LesiOnTime: Segmenting Small Lesions in High-Risk Breast Cancer
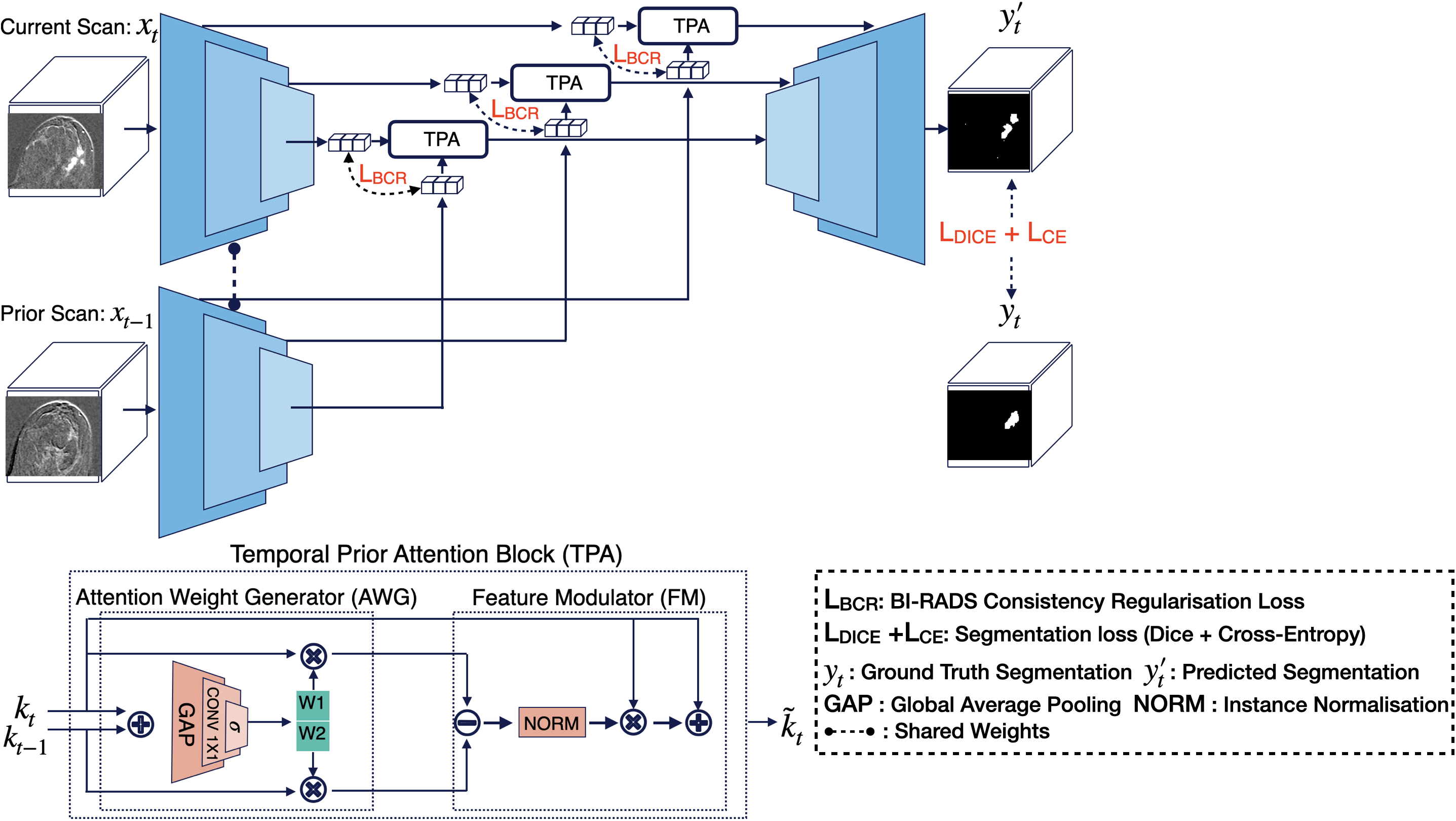
LesiOnTime is a novel 3D deep learning framework for longitudinal lesion segmentation that integrates temporal information from previous scans with clinical knowledge as a prior. Developed for high-risk breast cancer patients using longitudinal DCE-MRI, it introduces a Temporal Prior Attention (TPA) block that dynamically weights information across multiple scans, and a BI-RADS Consistency Regularization (BCR) loss that incorporates radiologists’ BI-RADS scores to guide the model. Explore the paper and code by Kamran Mohammed to learn more. It is part of EUCAIM and MALBACS.
Disentanglement of Biological and Technical Factors in Medical Images
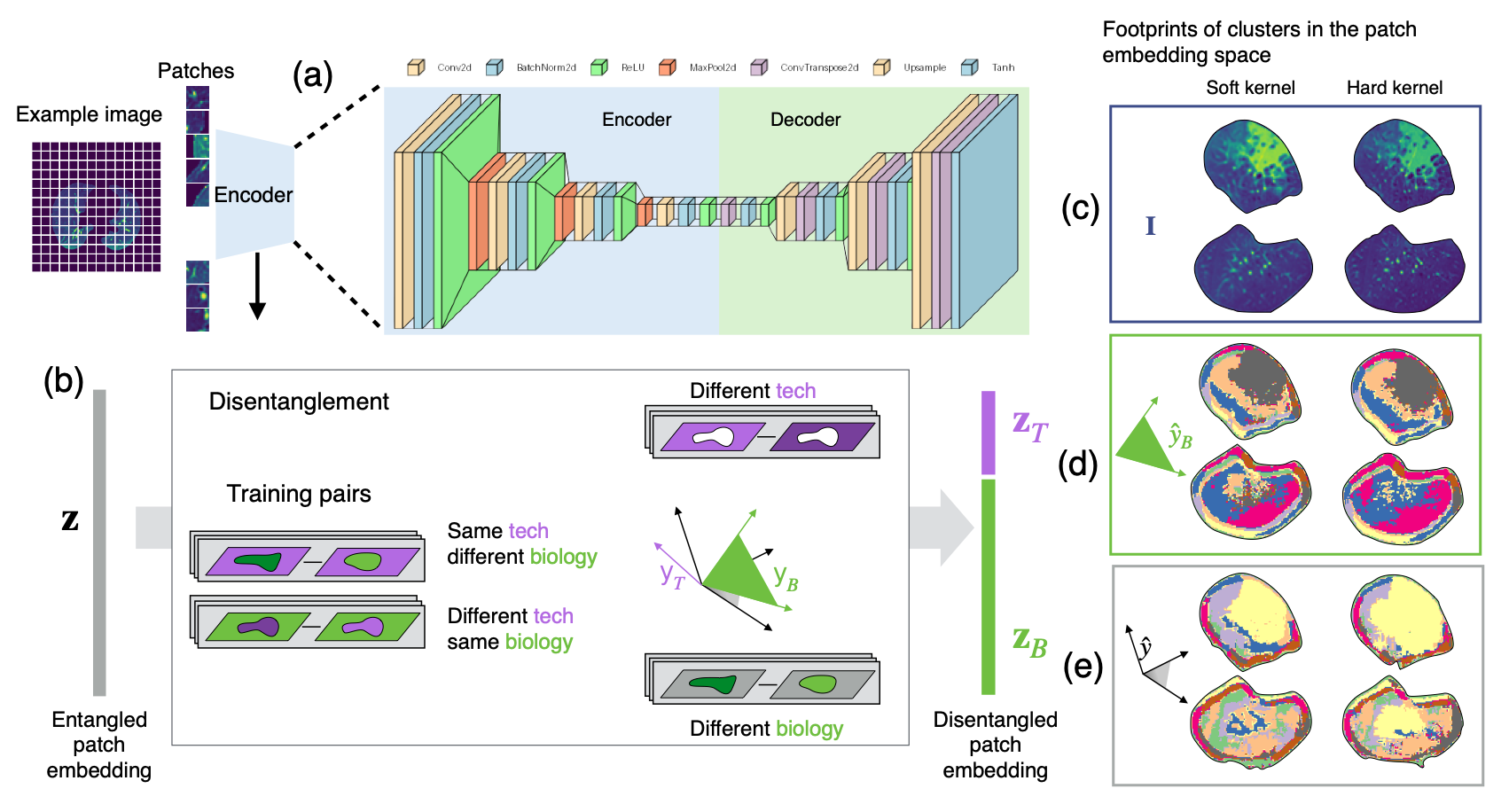
This research by Jeanny Pan and her collaborators introduces an approach that disentangles disease-related factors from scanner variability in CT images. By separating biological from technical factors, the method enables stable, label-free disease pattern discovery on external clinical datasets. This innovation drives biomarker discovery, strengthens survival prediction, and ensures reliable insights across multi-center routine imaging studies.
Learning with missing modalities, at train or test time.
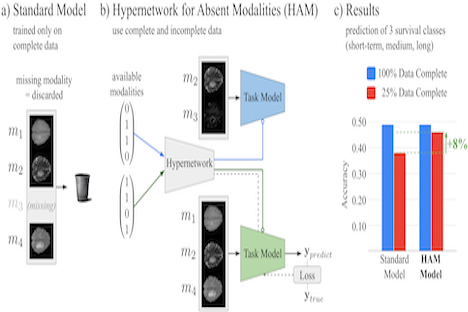
Christoph Fürböck propose a novel approach to cope with partially incomplete multi-modal data during training and test time. Introducing a hypernetwork that learns to generate a task model adapted to available modalities. During training it uses all available data and does not rely on an imputation model. During test time, it generates a task specific model that is optimized as a whole for the available set of modalities.
Unsupervised Anomaly Segmentation in Fine Visual Structure
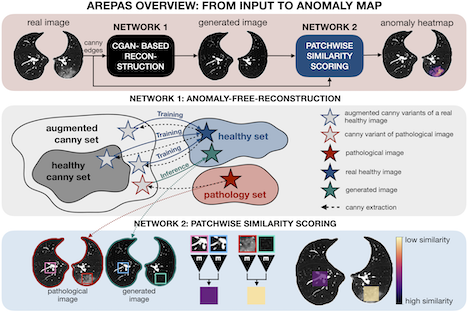
Normal fine-grained tissue variability such as present in pulmonary anatomy is a major challenge for existing generative Anomaly Detection (AD) methods. In his paper, Branko Mitic proposes a novel generative AD approach addressing this issue. It consists of an image-to-image translation for anomaly-free reconstruction and a subsequent patch similarity scoring between observed and generated image-pairs for precise anomaly localization. Branko and his team validate the new method on chest computed tomography (CT) scans for the detection and segmentation of infectious disease lesions.
Fetal Brain Atlas for Real Time Tissue Segmentation
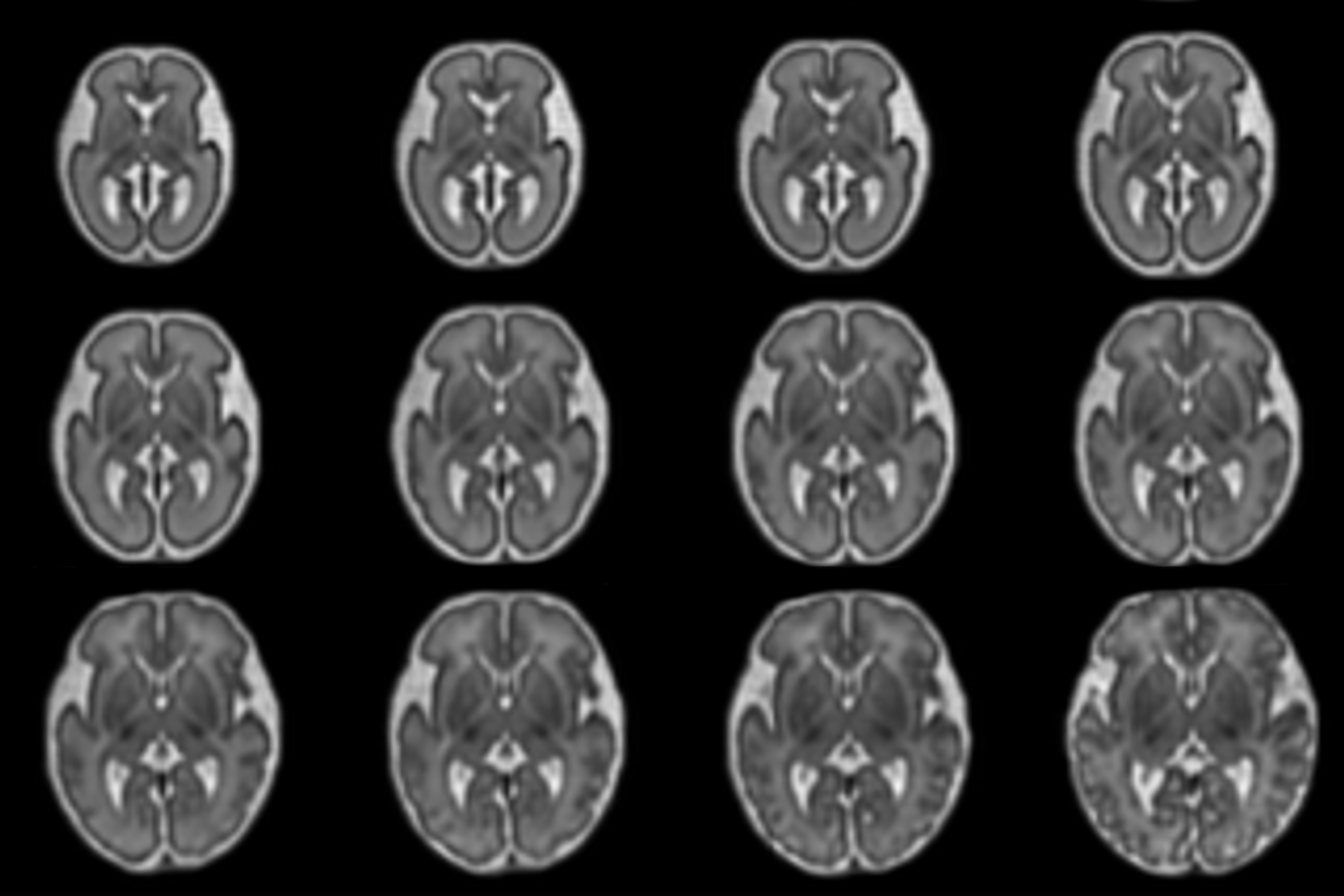
Johannes Tischer presents a registration-based framework for conditional atlas learning that captures healthy fetal brain development from 21 to 37 gestational weeks. By aligning the atlas to individual subjects, it enables automatic, real-time tissue segmentation that adapts to gestational age. The framework enhances clinical analysis and provides new insights into prenatal brain trajectories. Read more here.
Whole-Mount Sectioning Mosaicing (WMS) in Histopathology
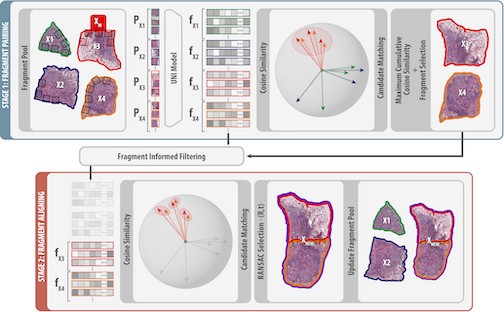
Stefan Brandstätter introduces SemanticStitcher, a method for automatically reconstructing whole-mount histopathology slides from fragmented tissue images. Unlike traditional boundary-based stitching, it leverages semantic features from visual foundation models to robustly align fragments despite distortions, staining inconsistencies, or missing regions. Evaluations across multiple datasets show that SemanticStitcher consistently outperforms state-of-the-art approaches, enabling more reliable large-scale histopathology analysis.
Mapping the Ganglionic Eminence in 3D: an automated tool to encode its growth trajectory in the fetal brain MRI.
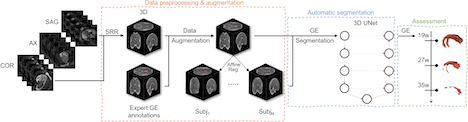
The ganglionic eminence (GE) is a transient fetal brain structure crucial for neural migration, yet automatic segmentation is hindered by small size, changing morphology, and image contrast inconsistency. Tomasso proposes a deep learning–based 3D UNet, trained with augmented fetal data generated by a registration-driven approach, to segment the GE from 19 to 38 gestational weeks. This approach enables to capture and analyse the GE growth trajectories in fetal brain MRI.
NeXtMarker – Fully Unsupervised and Interpretable Single-Cell Phenotyping in Multiplex Imaging
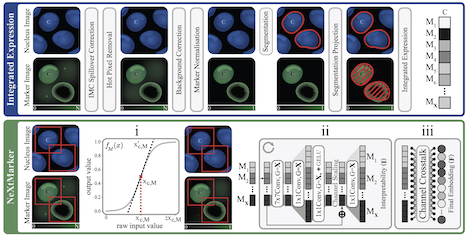
In this work Simon presents NeXtMarker, a contrastive learning framework for multiplex imaging that links marker expression with morphology in an interpretable feature space. It supports expert-driven phenotyping, matches or exceeds standard workflows, and generalizes effectively to new datasets. Across diverse data it can reliably distinguish cell types and highlight marker contributions, demonstrating its utility for robust, interpretable cell phenotyping.
