Research on anomaly detection spans a line of research aiming to extend the vocabulary of imaging markers beyond those we already know. We are developing several approaches to detect, segment and categorize anomalies.
Highlight Publications
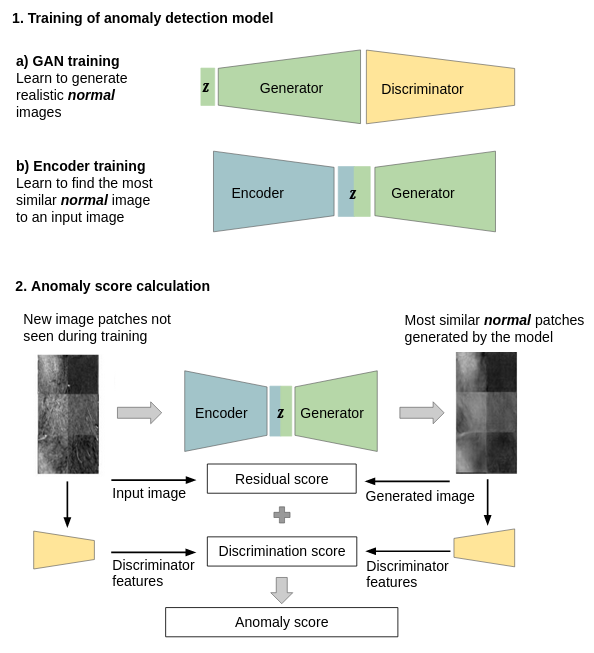
Generative Adversarial Network (GAN) Using a GAN for anomaly detection was initially inspired by an image completion approach. For anomaly detection the GAN learns the distribution of normal image examples and is used to to generate normal versions of new images that are checked for anomalies. Anoumalous images deviate more from the corresponding generated normal version than nomal images and can therfore be identified as such. The initial approach for anomaly detection devoped for anomalies in OCT images has been extended methodologically as well as to the detection of breast lesions in breast MRI (Schlegel et al, 2017, 2019, Burger et al, 2020, 2023). The code for GAN based anomaly detection is available: code releases
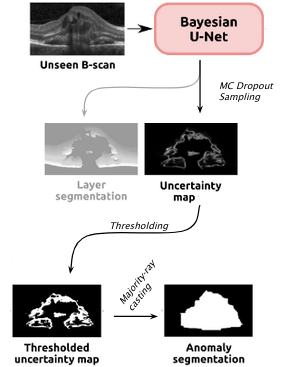
Epistemic Uncertainty Epistemic uncertainty was observed to increase when estimated on image samples whose appearance differ significantly from those on the training data. Thefore estimates from a Bayesian U-Net, that was trained for segmenting the anatomy of healthy subjects, were used as basis for anomaly detection. For this purpose obtained uncertainty maps were first binarized and then refined to yield a segmentation of the anomaly. The refinement is performed through a "majority-ray casting" technique, that makes use of the visual appearance of the retina in OCT. (Seeböck et al, 2019)
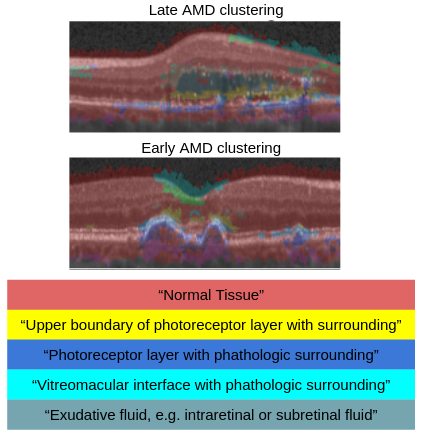
Identification of Disease Marker Candidates in OCT A multi-scale deep denoising autoencoder was trained on healthy images, and a one-class support vector machine identified anomalies in new data. Clustering in the anomalies identified stable categories as marker candidates occurring frequently in the data. Using these markers to classify healthy-, early AMD- and late AMD cases yields an accuracy of 81.40%. In a second binary classification experiment on a publicly available data set (healthy vs. intermediate AMD) the model achieves an AUC of 0.944. A careful qualitative analysis of the identified data driven markers reveals how their quantifiable occurrence aligns with our current understanding of disease course, in early- and late age-related macular degeneration (AMD) patients. (Seeböck et al, 2019)

Phenotyping of lung CTs A key question in learning from clinical routine imaging data is whether we can identify coherent patterns that re-occur across a population, and at the same time are linked to clinically relevant patientparameters. Here, we present a feature learning and clustering approach that groups 3D imaging data based on visual features at corresponding anatomical regions extracted from clinical routine imaging data without any supervision. (Hofmanninger et al, 2016)
Publications
- Burger B, Bernathova M, Helbich T, Singer C F, Langs G: Deep learning for predicting future lesion emergence in high-risk breast MRI screening: a feasibility study, European Readiology Experimental 7, 32, (2023)
- Burger B, Bernathova M, Helbich T, Singer C F, Langs G: AI-based prediction of lesion occurrence in high-risk women based on anomalies detected in follow-up examinations, 15th International Workshop on Breast Imaging (IWBI2020). International Society for Optics and Photonics, p 115130P
- Seeböck P., Orlando J.I., Schlegl T., Waldstein S., Bogunovic H., Klimscha S., Langs G., Schmidt-Erfurth U: Exploiting Epistemic Uncertainty of Anatomy Segmentation for Anomaly Detection in Retinal OCT, IEEE Transactions on Medical Imaging, (2019)
- Matthias Perkonigg, Daniel Sobotka, Ahmed Ba-Ssalamah, Georg Langs: Unsupervised deep clustering for predictive texture pattern discovery in medical images. Proc. Med-NeurIPS (2019), arXiv
- Thomas Schlegl, Philipp Seeböck, Sebastian M. Waldstein, Georg Langs*, Ursula Schmidt-Erfurth: f-AnoGAN: Fast Unsupervised Anomaly Detection with Generative Adversarial Networks, Medical Image Analysis, (2019)
- Seeböck P., Waldstein S., Klimscha S., Bogunovic H., Schlegl T., Gerendas B. S., Donner R., Schmidt-Erfurth U., Langs G: Unsupervised Identification of Disease Marker Candidates in Retinal OCT Imaging Data, IEEE Transactions on Medical Imaging, (2019)
- T Schlegl, P , S M. Waldstein, U Schmidt-Erfurth, G Langs: Unsupervised Anomaly Detection with Generative Adversarial Networks to Guide Marker Discovery, International Conference on Information Processing in Medical Imaging, pp. 146-157. Springer, Cham, 2017. [arxiv]
- Seeböck, P., Donner, R., Schlegl, T., & Langs, G: Unsupervised Learning for Image Category Detection. Proceedings of the 22nd Computer Vision Winter Workshop. (2017, Best Paper Award)
- J. Hofmanninger, M. Krenn, M. Holzer, T. Schlegl, H. Prosch, G. Langs: Unsupervised Identification of Clinically Relevant Clusters in Routine Imaging Data. Published in Proceedings of MICCAI (2016).
- Philipp Seeböck, Sebastian Waldstein, Sophie Klimscha, Bianca S. Gerendas, René Donner, Thomas Schlegl, Ursula Schmidt-Erfurth, Georg Langs. Identifying and Categorizing Anomalies in Retinal Imaging Data. NIPS MLHC (2016)
Images: MUW/Burger, Seeböck, Seeböck, Langs
